Researchers from the Defense Threat Reduction Agency’s Joint Science and Technology Office (JSTO) are at the forefront of developing a paper-based diagnostic device to detect viral threats, such as Zika. Managed by JSTO’s Dr. Ilya Elashvili, the project is led by Dr. James J. Collins and his team from the Massachusetts Institute of Technology.
In a paper recently published in the journal Cell, the team details how they created a portable diagnostic platform that allows rapid, specific and low-cost detection of the Zika virus at clinically relevant concentrations. To achieve this outcome, the team merged two biotechnologies, which they previously developed.
Toehold Switches
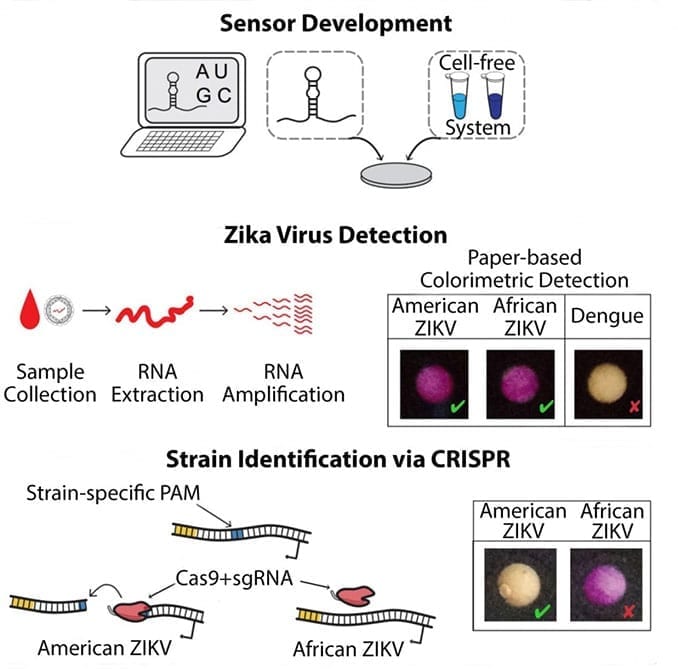
The first of the two biotechnologies uses a riboregulator concept to construct programmable ribonucleic acid (RNA) sensors called toehold switches, which can be rationally designed to bind and sense virtually any RNA sequence. This sensing portion of the toehold is placed upstream of a ribosome binding site (RBS) to create a RBS-locking hairpin structure, which block RNA binding to ribosome and thereby prevents the translation of a downstream reporter protein.
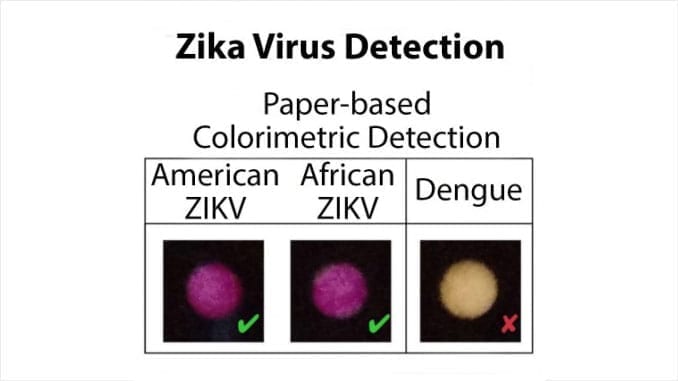
When targeted, “trigger” RNA binds the toehold, peeling the hairpin and opening the blockade, allowing the RNA to bind to the ribosome and subsequent translation of the reporter protein (LacZ enzyme). This converts a yellow substrate to a purple product, indicating the presence of the virus.
Protein Expression Platform
The second technology involves a paper-based, freeze-dried, cell-free protein expression platform that allows for the deployment of these toehold switch sensors outside of a research laboratory by providing a sterile method for storage and distribution of genetic circuits at room temperature.
In order to achieve the sensitivity needed in the diagnostic tests, the researchers employed widely used nucleic acid sequence-based amplification (NASBA) to isothermally amplify extracted RNA. As a result, the researchers were able to develop a low-cost diagnostic platform for the detection of clinically relevant (low femtomolar) concentrations of Zika virus from infected monkey plasma.
Notably, there was no significant detection of the closely related Dengue virus.
CRISPR-Cas9 Genotyping
By employing a Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9-based module that can cut preprogrammed sequences in NASBA amplified product and thereby prevent reporting the selected target sequences, the researchers were able to discriminate different genotypes of Zika virus (i.e., American Zika virus from African Zika virus).
This diagnostic tool can be stored for more than a year at room temperature and does not require highly skilled personnel or expensive equipment to operate. This makes it suitable for current or emerging viral threats for deployed warfighters in low-resource areas, where detection and containment are critically needed.
Read more: Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components
Article based on original by Dr. Ilya Elashvili, DTRA. Edited for context and format by Global Biodefense.