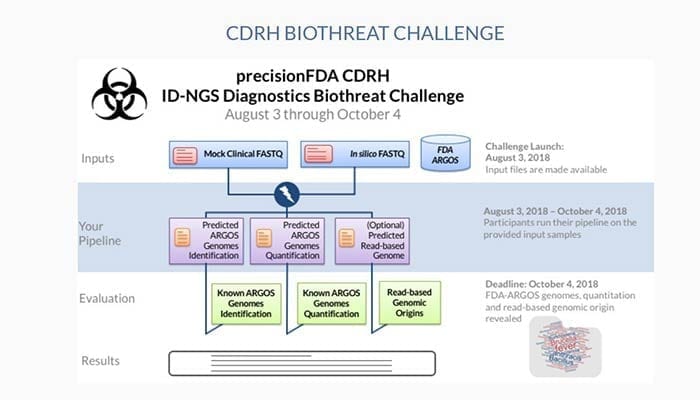
To encourage the development and improvement of Infectious Diseases Next-Generation Sequencing (ID-NGS) analytical methods, precisionFDA – the community platform for NGS assay evaluation and regulatory science exploration – has launched the precisionFDA CDRH Infectious Disease NGS Diagnostics Biothreat Challenge.
Professional and citizen scientists are invited to test their bioinformatics skills and software tools in a challenge to identify pathogens from the FDA-ARGOS database within host samples using NGS short-read data.
The focus of this challenge is to enable tool developers to test their algorithms on blinded mock-clinical and in silico metagenomics samples using provided regulatory-grade reference genomes from the FDA-ARGOS database. This will enable the community to look at bioinformatics pipeline performance using a fixed reference genome data standard. The challenge will help familiarize precisionFDA users with the agency’s innovative FDA-ARGOS database resource (www.fda.gov/argos).
The challenge is open now through October 4, 2018.
About FDA-ARGOS
A publicly available database for Reference Grade microbial Sequences, called FDA-ARGOS, was established by the FDA and collaboraters in 2014. With funding support from FDA’s Office of Counterterrorism and Emerging Threats (OCET) and DoD, the FDA-ARGOS team are initially collecting and sequencing 2000 microbes that include biothreat microorganisms, common clinical pathogens and closely related species.
Currently, FDA-ARGOS microbial genomes are typically generated in 3 phases:
- Phase 1 entails collection of a previously identified microbe and nucleic acid extraction
- Phase 2, the microbial nucleic acids are sequenced and de novo assembled using Illumina and Pac Biosequencing platforms at the Institute for Genome Sciences at the University of Maryland (UMD-IGS)
- Phase 3, the assembled genomes are vetted by an ID-NGS subject matter expert working group consisting of FDA personnel and collaborators and the data are deposited in NCBI databases
Manufacturers who develop sequence-based test to identify infectious agents and/or to detect resistance or virulence markers can use FDA-ARGOS to advance their development programs and to support the regulatory science review of such test. For example, FDA-ARGOS can be used as a tool for in-silico (computer simulation) data analysis.
Additional Resources
- In Vitro Diagnostics
- Access the FDA-ARGOS Database @ NCBI
- Guidance Document: Infectious Disease Next Generation Sequencing Based Diagnostic Devices: Microbial Identification and Detection of Antimicrobial Resistance and Virulence Markers – Draft Guidance for Industry and Food and Drug Administration Staff
Source: Food and Drug Administration