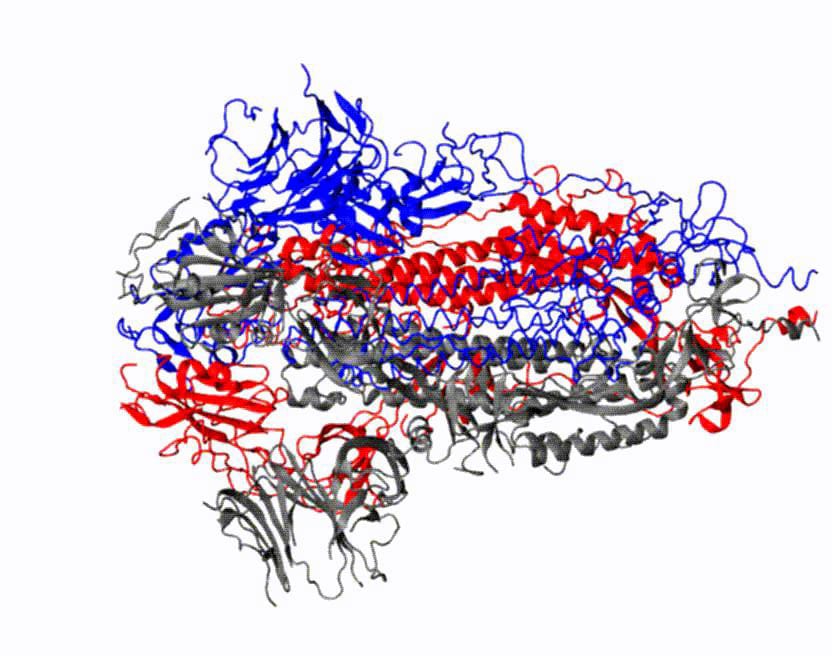
Research will show how the virus binds to human cell receptors and will aid the development of vaccines and therapeutics
Computational chemist Mahmoud Moradi will develop enhanced, 3D simulations of the molecular dynamics of coronavirus spike glycoproteins to gain better understanding of how the virus binds to human cells.
Moradi’s work is part of the COVID-19 High Performance Computing Consortium, a collaboration of government, industry and academic partners focused on computing resources for COVID-19 research. Spearheaded by the White House Office of Science and Technology Policy (OSTP), the U.S. Department of Energy and IBM, the consortium volunteers free compute time and resources on some of the world’s most powerful supercomputers.
Mapping how these proteins undergo conformational changes to bind to host cell receptors is critical to the development of coronavirus vaccines and therapeutics. Simulations are especially important because a framework for drug design will require dynamic, three-dimensional visualizations of cell structures and behavior, rather than a static picture.
“As with other viruses, a crucial step in the coronavirus infection process is viral entry,” said Moradi, assistant professor in the J. William Fulbright College of Arts and Sciences. “With coronaviruses, we know these spike glycoproteins mediate entry into the human cell. Both SARS-CoV-2, the cause of COVID-19, and SARS-CoV, the cause of the 2002-2003 SARS epidemic, have spike proteins that attach to the same receptor in human cells.”
To perform the simulations, Moradi has been granted access to Frontera, a National Science Foundation-sponsored supercomputer housed at the University of Texas at Austin. Frontera is the largest supercomputer on any university campus.
Moradi’s project benefits from several recent, high-resolution 3D models of the coronavirus spike proteins. These models can be used as initial structures to begin simulations that will enable analysis of the detailed mechanisms of the proteins and their behavior upon viral entry. Enhanced, detailed simulations of such molecular dynamics will provide a complete picture of proteins’ structural changes, as well as how they bind to angiotensin-converting enzyme 2, the specific human cell receptor.
Moradi’s research lies at the intersection of biology, physics, chemistry, mathematics, statistics and computer science. His biomolecular simulations and computational theories explain how proteins, the workhorse molecules of cells, function at the molecular level. His work improves geometric models to describe how proteins change their shape and how these changes affect a protein’s behavior. In February, he received a $650,000 National Science Foundation Faculty Early Career Development award for this work.