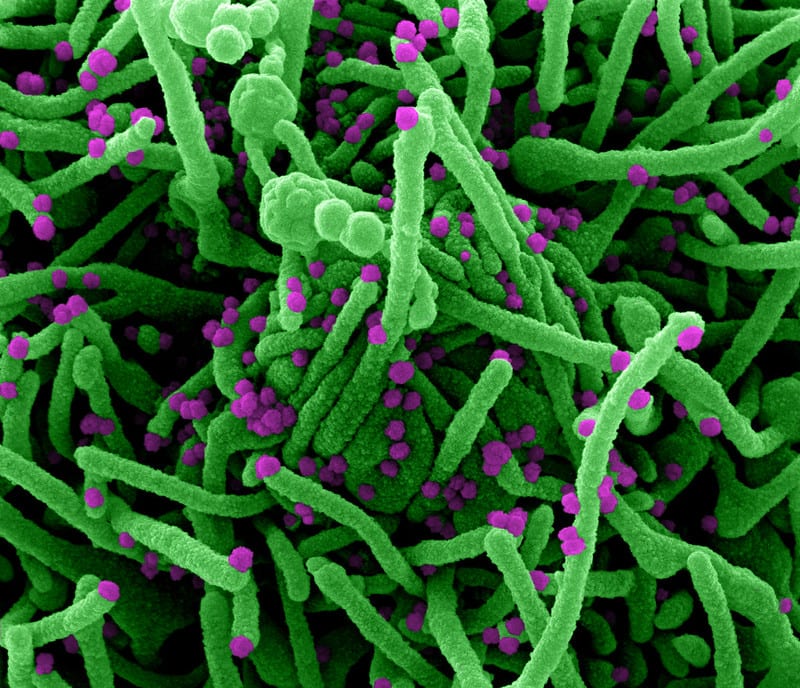
Scientists in Brazil recently reported that two people were simultaneously infected with two different variants of SARS-CoV-2, the virus that causes COVID-19. This co-infection seemed to have no effect on the severity of patients’ illness, and both recovered without needing to be hospitalised.
Although this is one of the few such cases recorded with SARS-CoV-2 – and the study is yet to be published in a scientific journal – scientists have observed infections with multiple strains with other respiratory viruses, such as influenza. This has raised questions about how these viruses may interact in an infected person, and what it could mean for generating new variants.
Viruses are masters of evolution, constantly mutating and creating new variants with every cycle of replication. Selective pressures in the host, such as our immune response, also drive these adaptations. Most of these mutations won’t have a significant effect on the virus. But ones that give an advantage to the virus – for example, by increasing its ability to replicate or evade the immune system – are cause for concern and need to be closely monitored.
The occurrence of these mutations is down to the error-prone replication machinery that viruses use. RNA viruses, such as influenza and hepatitis C, generate a relatively large number of errors each time they replicate. This creates a “quasi-species” of the virus population, rather like a swarm of viruses, each with related but non-identical sequences. Interactions with the host cells and immune system determine the relative frequencies of the individual variants, and these coexisting variants may affect how the disease progresses or how well treatments work.
Compared with other RNA viruses, coronaviruses have lower mutation rates. This is because they are equipped with a proofreading mechanism that can correct some of the errors that occur during replication. Still, there is evidence of viral genetic diversity in patients infected with SARS-CoV-2.
The detection of multiple variants in a person could be the result of co-infection by the different variants, or the generation of mutations within the patient after the initial infection. One way to discriminate these two scenarios is by comparing the sequences of the variants circulating in the population with those in the patient. In the Brazilian study mentioned above, the variants identified corresponded to different lineages that had been previously detected in the population, implying co-infection by the two variants.
Mixing It All Up
This co-infection has opened concerns of SARS-CoV-2 acquiring new mutations even more rapidly. This is because coronaviruses can also undergo large changes in their genetic sequence by a process called recombination. When two viruses infect the same cell, they can swap large parts of their genomes with each other and create completely new sequences.
This is a known phenomenon in RNA viruses. New variants of influenza are generated by a similar mechanism called “reassortment”. The genome of influenza virus, unlike coronavirus, comprises eight segments or strands of RNA. When two viruses infect the same cell, these segments mix and match to produce viruses with a new combination of genes. Interestingly, pigs can be infected with different strains of influenza viruses, and have been referred to as “mixing vessels” that shuffle them into new strains. The 2009 H1N1 pandemic virus emerged from a reassortment of a human, avian, and two swine influenza viruses.
With coronaviruses, which only contain one RNA strand in each virus particle, recombination can only occur between RNA strands derived from one or more viruses in the same cell. Evidence of recombination has been found both in the laboratory and in a patient infected with SARS-CoV-2, suggesting that this could drive the generation of new variants. In fact, the ability of SARS-CoV-2 to infect human cells is proposed to have developed via recombination of the spike protein between closely related animal coronaviruses.
It is important to note that this requires the two viruses to infect the same cell. Even if a person is infected with several variants, if they replicate in different parts of the body, they will not interact with each other. Indeed, this was seen in patients, where different quasi-species of coronaviruses were found in the upper and lower respiratory tracts, suggesting that viruses in these sites were not directly mixing with each other.
The evidence so far does not suggest that infection with more than one variant leads to more severe disease. And although possible, very few cases of co-infection have been reported. More than 90% of the infections in the UK currently are by B117 – the so-called Kent variant. With such a high prevalence of one variant in the population, co-infections are not likely to occur. Still, monitoring this landscape allows scientists to track the emergence of these new variants of concern and understand and respond to any changes in their transmission or vaccine efficacy.
ABOUT THE AUTHOR
Maitreyi Shivkumar is a Senior Lecturer in Molecular Biology in the Leicester School of Pharmacy at De Montfort University. Her research interests are in understanding host-pathogen interactions, with a focus on virology. She completed her PhD at the University of Cambridge on host entry and dissemination of herpesviruses, and was a postdoctoral researcher at University College London studying HIV-1 spread, before joining De Montfort University in 2019.
This article courtesy of The Conversation.