There are only two days left to complete the CFSAN Pathogen Detection Challenge.
Sponsored by the U.S. Food and Drug Administration (FDA), Center for Food Safety and Applied Nutrition (CFSAN), the challenge calls on the food safety and infectious diseases communities to help improve bioinformatics pipelines for detecting pathogens in samples sequenced using metagenomics.
As the food safety community moves to metagenomic sequencing, bioinformatics algorithms must be developed to detect pathogens amongst a mix of genetic material sequenced directly from a sample. Within bioinformatics and genomics, crowdsourcing has a history of improving expertly developed algorithms.
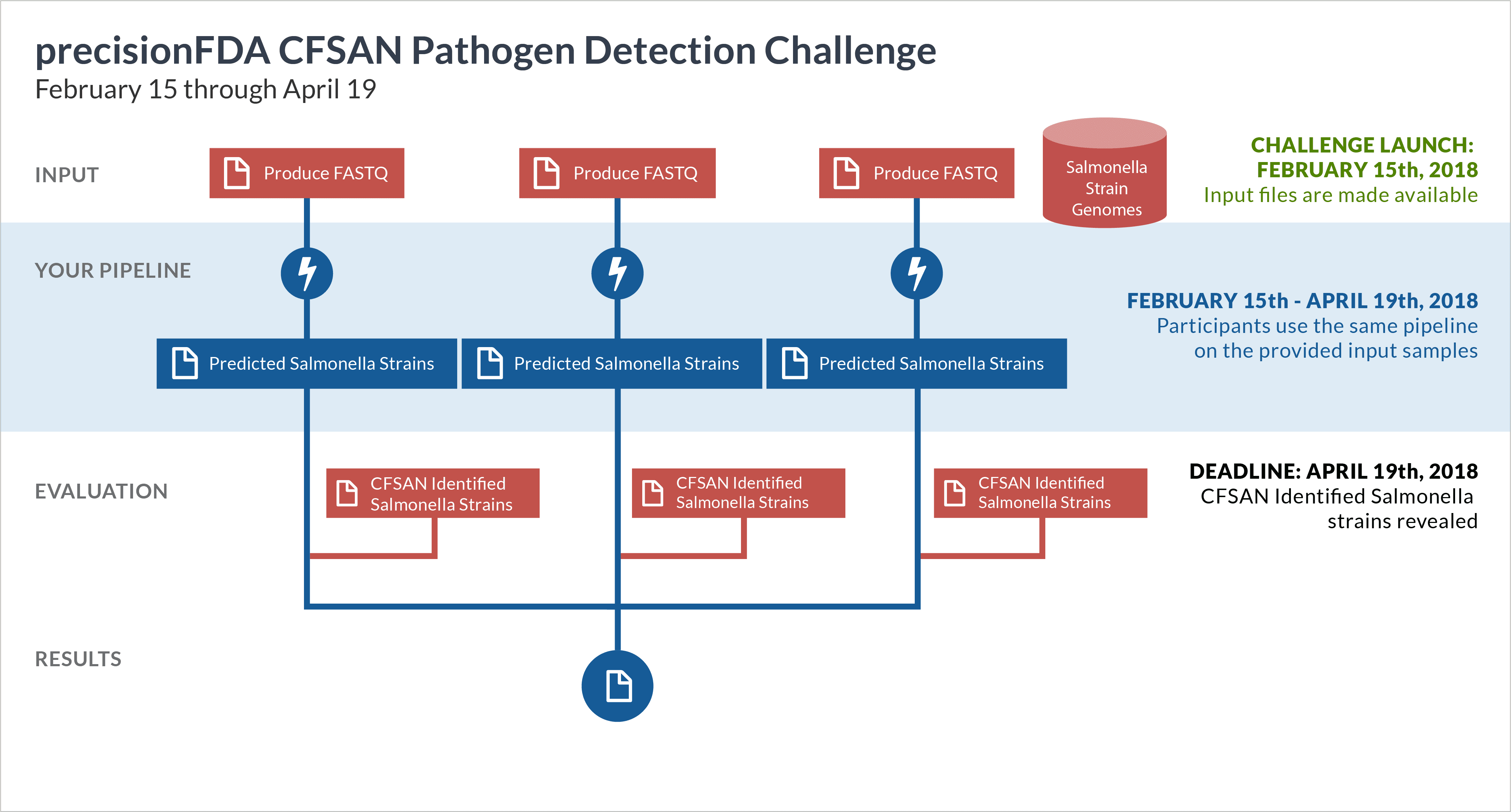
In this challenge, participants are asked to develop and use bioinformatics pipelines to identify the types and specific Salmonella strains in each of several metagenomics samples. This type of technology will expedite determining the source of foodborne illness.
In the U.S. alone, one in six individuals, an estimated 48 million people, fall prey to foodborne illness, resulting in 128,000 hospitalizations and 3,000 deaths per year. Economic burdens are estimated cumulatively at $152 billion dollars annually, including $39 billion due to contamination of fresh and processed produce. One longstanding problem is the ability to rapidly identify the food-source associated with the outbreak being investigated.
The faster an outbreak is identified and the increased certainty that a given source and patients are linked, the faster the outbreak can be stopped, limiting morbidity and mortality.
Submit your results today at http://go.usa.gov/xnHa6
Source: FDA, adapted.