 A new study by researchers at the Georgia Institute of Technology suggests that the number of unique pocket sites where small molecule pharmaceutical compounds can bind to proteins is surprisingly small, meaning drug side effects may be impossible to avoid.
A new study by researchers at the Georgia Institute of Technology suggests that the number of unique pocket sites where small molecule pharmaceutical compounds can bind to proteins is surprisingly small, meaning drug side effects may be impossible to avoid.
Studying a set of artificial proteins and comparing them to natural proteins, the team concluded that there might be no more than about 500 unique protein pocket configurations that serve as binding sites for small molecule ligands. Therefore, the likelihood that a molecule intended for one protein target will also bind with an unintended target is significant, said Jeffrey Skolnick a professor in the School of Biology at Georgia Tech.
“Our study provides a rationalization for why a lot of drugs have significant side effects – because that is intrinsic to the process,” said Skolnick. “There are only a relatively small number of different ligand binding pockets. The likelihood of having geometry in an amino acid composition that will bind the same ligand turns out to be much higher than anyone would have anticipated. This means that the idea that a small molecule could have just one protein target can’t be supported.”
Binding pockets on proteins are formed by the underlying secondary structure of the amino acids, which is directed by hydrogen bonding in the chemistry. That allows formation of similar pockets on many different proteins, even those that are not directly related to one another.
“You could have the same or very similar pockets on the same protein, the same pockets on similar proteins, the same pockets on completely dissimilar proteins that have no evolutionary relationship. In proteins that are related evolutionarily or that have similar structures, you could have very dissimilar pockets,” said Skolnick, who is also a Georgia Research Alliance Eminent Scholar. “This helps explain why we see unintended effects of drugs, and opens up a new paradigm for how one has to think about discovering drugs.
The implications of this “biochemical noise” for the drug discovery process could be significant. To counter the impact of unintended effects, drug developers will need to know more about the available pockets so they can avoid affecting binding locations that are also located on proteins critical to life processes. If the inevitable unintended binding takes place on less critical proteins, the side effects may be less severe.
In addition, drug development could also move to a higher level, examining the switches that modulate the activity of proteins beyond binding sites. That may require a different approach to drug development.
“The strategy for minimizing side effects and maximizing positive effects may have to operate at a higher level,” he said. “You are never going to be able to design unintended binding effects away. But you can minimize the undesirable effects to some extent.”
Read more about the research at PNAS: Interplay of physics and evolution in the likely origin of protein biochemical function.
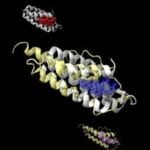
Image shows a naturally heme-bound (blue) protein structure (white, pocket in red), superimposed onto an artificial protein structure (yellow, pocket in purple) that has both a similar protein fold and a similar pocket. The pocket-like features of proteins can be artificially generated in stable proteins without invoking evolutionary selection. (Image courtesy of Mu Gao, Georgia Tech).
Source: Georgia Institute of Technology


