A research team from Vanderbilt University Medical Center is the first to decipher the 3-D structure of a protein that confers antibiotic resistance from one of the most worrisome disease agents: methicillin-resistant Staphylococcus aureus (MRSA). The Vanderbilt team’s findings may be an important step in combating the MRSA public health threat over the next 5 to 10 years.
By deciphering the shape of a key S. aureus protein — an enzyme called FosB that inactivates an antibiotic called fosfomycin — the researchers have set the stage to devising a therapeutic method to inhibit FosB and hence improve the efficacy of the antibiotic.
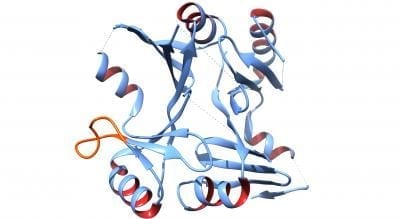
“Our hope is that now that we know the 3-D shape and overall function of the FosB protein, we will be able to design inhibitors of FosB that will enable fosfomycin to function appropriately as an antibiotic,” said Matthew K. Thompson, a postdoctoral researcher on the team. “When we can successfully do that, we may very well be able to combat S. aureus infections with fosfomycin.”
Identifying the FosB protein’s three-dimensional structure helps scientists understand that protein’s particular function. In particular, the new structural images of FosB, obtained by a technique called X-ray crystallography, provide insight into the functional role of a particular part of the protein’s shape called a binding loop. It appears to function like a door that opens and closes to allow the antibiotic to enter the active site of FosB.
In addition to providing new insight on the function of this S. aureus protein, the research has also produced new evidence for the role zinc might play in inhibiting FosB. This could impact restoring the efficacy of fosfomycin, leading to treatment for a variety of multi-drug-resistant pathogens.
The team presented its work this week at the 58th Annual Meeting of the Biophysical Society: Structure and Function of the Genomically-encoded Fosfomycin Resistance Enzyme, FOSB, from Staphylococcus aureus.

