 Nephele, sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), assists microbiome researchers by providing a variety of standardized analysis pipelines assembled using tools that include QIIME, mothur, and bioBakery; and then download visualizations and easy-to-interpret results.
Nephele, sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), assists microbiome researchers by providing a variety of standardized analysis pipelines assembled using tools that include QIIME, mothur, and bioBakery; and then download visualizations and easy-to-interpret results.
The NIAID Office of Cyber Infrastructure and Computational Biology (OCICB) has sponsored public use of Nephele for a special promotional period, in an effort to address current challenges surrounding biomedical big data and to make digital research objects more accessible through the use of cloud computing.
Nephele is targeted to researchers, sequencing facilities, students, and citizen scientists who would like to perform standardized microbiome analyses on amplicon or shotgun sequencing data.
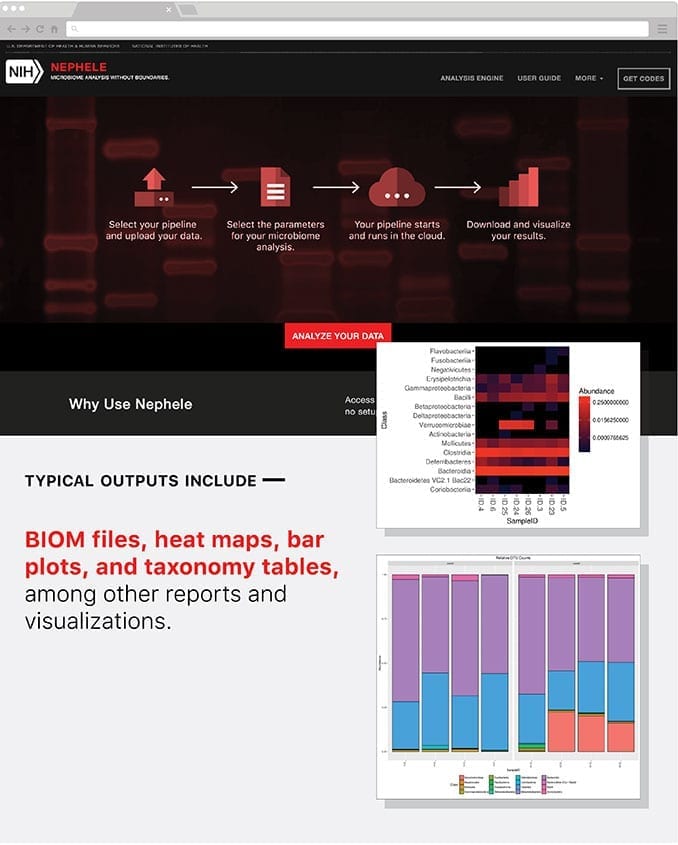
Registration is free and once sent an access code, users simply choose the appropriate pipeline and data type for 16S, 18S, ITS, or WGS analysis, provide their data files through upload or a URL link, optionally set a few processing parameters, and click submit.
Once processing is complete, results can be interpreted with help from Nephele’s tutorial videos or Nephele’s Pipeline Output Guide. Typical outputs include BIOM files, heat maps, bar plots, and taxonomy tables, among other reports and visualizations.