How can we battle these infectious agents such as Ebola virus disease (EVD) that reproduce by hijacking cells and reprogramming them into virus-replicating machines?
A research team led by Professor Juan Perilla at the University of Delaware are using supercomputers to simulate the inner workings of Ebola, observing the way molecules move, atom by atom, to carry out their functions. In a new paper, the researchers detail structural features of the virus’s coiled protein shell, or nucleocapsid, that may be promising therapeutic targets, more easily destabilized and knocked out by an antiviral treatment.
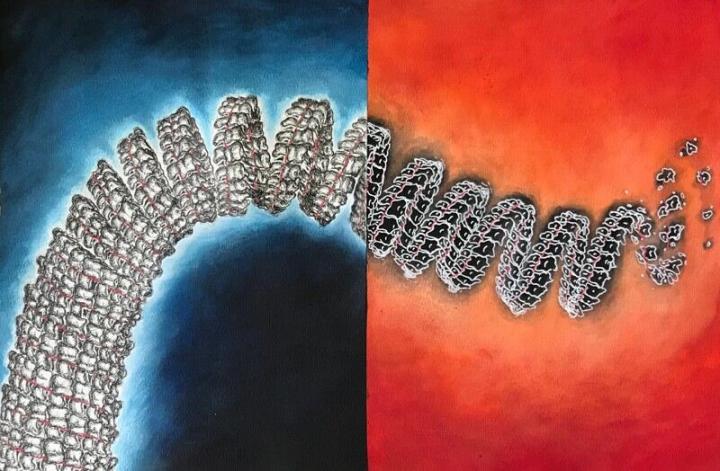
“The Ebola nucleocapsid looks like a Slinky walking spring, whose neighboring rings are connected,” Perilla said. “We tried to find what factors control the stability of this spring in our computer simulations.”
The life cycle of Ebola is highly dependent on this coiled nucleocapsid, which surrounds the virus’s genetic material consisting of a single strand of ribonucleic acid (ssRNA). Nucleoproteins protect this RNA from being recognized by cellular defense mechanisms. Through interactions with different viral proteins, such as VP24 and VP30, these nucleoproteins form a minimal functional unit — a copy machine — for viral transcription and replication.
While nucleoproteins are important to the nucleocapsid’s stability, the team’s most surprising finding, Perilla said, is that in the absence of single-stranded RNA, the nucleocapsid quickly becomes disordered. But RNA alone is not sufficient to stabilize it. The team also observed charged ions binding to the nucleocapsid, which may reveal where other important cellular factors bind and stabilize the structure during the virus’s life cycle.
Perilla compared the team’s work to a search for molecular “knobs” that control the nucleocapsid’s stability like volume control knobs that can be turned up to hinder virus replication.
The UD team built two molecular dynamics systems of the Ebola nucleocapsid for their study. One included single-stranded RNA; the other contained only the nucleoprotein. The systems were then simulated using the Texas Advanced Computing Center’s Frontera supercomputer – the largest academic supercomputer in the world. The simulations took about two months to complete.
While a vaccine exists for Ebola, it must be kept extremely cold, which is difficult in remote African regions where outbreaks have occurred. Will the team’s work help advance new treatments?
“As basic scientists we are excited to understand the fundamental principles of Ebola,” Perilla said. “The nucleocapsid is the most abundant protein in the virus and it’s highly immunogenic — able to produce an immune response. Thus, our new findings may facilitate the development of new antiviral treatments.”
Currently, Perilla and Jodi Hadden-Perilla are using supercomputer simulations to study the novel coronavirus that causes COVID-19. Although the structures of the nucleocapsid in Ebola and COVID-19 share some similarities — both are rod-like helical protofilaments and both are involved in the replication, transcription and packing of viral genomes — that is where the similarities end.
“We now are refining the methodology we used for Ebola to examine SARS-CoV-2,” Perilla said.
Molecular determinants of Ebola nucleocapsid stability from molecular dynamics simulations. The Journal of Chemical Physics, 20 October 2020.