Antibiotic resistance – the ability of harmful bacteria to survive treatment by antibiotics – is a growing threat. It is making it harder to treat life-threatening infections, including tuberculosis, MRSA, and gonorrhoea – and increasing the risks of even minor surgery.
In order to solve antibiotic resistance, one thing researchers first need to understand is how to stop resistance from happening to begin with. A recent study I conducted with colleagues at the University of Oxford has helped increase that understanding by showing bacteria can cleverly rearrange their genetics in order to evade the effects of an antibiotic.
Bacteria have multiple ways of evolving resistance. They can mutate to prevent antibiotics from targeting them, which can be done by modifying the proteins within the cell where antibiotics act. They can also acquire genes that help them produce antibiotic-destroying molecules, called enzymes.
However, all these strategies carry a cost for resistant bacteria. Producing resistance enzymes requires a lot of energy. Modified proteins also cannot perform as effectively as before. Both these factors severely hamper bacteria, and make them replicate slower when antibiotics aren’t present. This leads resistant bacteria to lose the competition against other bacteria for precious nutrients and resources, threatening their survival.
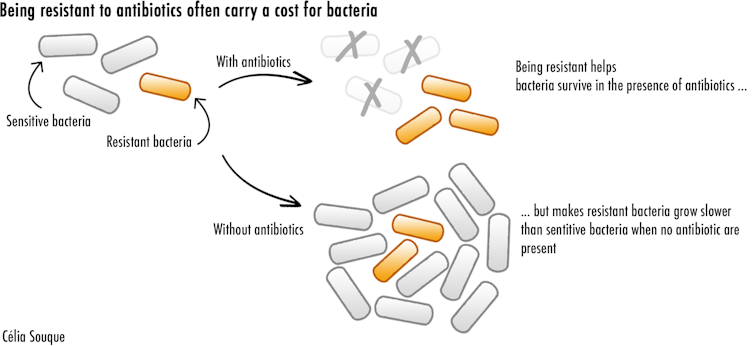
But resistant bacteria have found a way to become resistant to antibiotics while limiting the costs associated with it. My recent study showed how one such mechanism, involving something known as an integron, provides bacteria with an incredible potential to acquire high levels of resistance while reducing its energy cost. This makes it easier for antibiotic resistant bacteria to survive – and thrive.
Integrons are bits of DNA, unique to bacteria, that allow bacteria to stockpile genes they acquire from other resistant bacteria. These resistance genes are lined up in the bacteria genome one after the other forming “arrays”. The position of the genes in the array has a big impact on the bacteria’s resistance levels.
Genes that are present toward the start of the array are heavily expressed (meaning they’re actively being used) and provide high levels of resistance. Genes at the back are kept silent and can be conserved at low cost, reducing their impact on the bacteria.
On top of this, integrons come with a fantastic trick: an enzyme, called integrase, that allows bacteria to cut off and move genes in the array when the bacteria are in danger. The integrase is thought to provide bacteria with the ability to “shuffle” the order of their genes, letting bacteria modulate their resistance levels on demand. Our study was the first to test this hypothesis.
To see how useful integrons can be for bacteria, we built custom integrons in the lab which contained a relevant resistance gene in last position. Some were made to have a dysfunctioning integrase enzyme, which would prevent them from being able to move their genes around. This allowed us to measure the impact of gene shuffling on antibiotic resistance.
We then used an approach called experimental evolution where we challenged bacteria with increasing doses of antibiotics and observed how long they survived. This technique allowed us to directly measure how good bacteria are at evolving resistance.
We showed that the bacteria that could shuffle their genes survived longer and evolved resistance more frequently than the ones that couldn’t. This shows how integrons can help bacteria evolve high levels of antibiotic resistance in response to treatment with antibiotics.
Interestingly, this shuffling was often linked with loss of the other resistance genes present in the bacteria. By shuffling genes around to become resistant against our chosen antibiotic, bacteria lost some of their other resistance genes in the process – again becoming susceptible to these other antibiotics.
New Strategies
The results from our study provide potential strategies to counteract integrons and their role in evolving resistance. For example, antibiotics could be combined with drugs that can inhibit the enzyme integrase to reduce gene shuffling. Drugs that stop the bacteria’s “SOS response” – the bacteria’s last resort reaction to antibiotics – would also limit integron shuffling as well. So called “anti-evolution” drugs, which do not kill bacteria directly but help prevent the evolution of resistance, are currently an active area of research.
Another alternative would be to exploit the integron shuffling to promote the loss of resistance genes by cycling through different antibiotics. This would steer the evolution of bacteria in a way that makes them sensitive to previously unusable antibiotics.
Integrons first evolved millions of years ago. But now they’ve found themselves to be a uniquely suited mechanism for bacteria to adapt to the use of antibiotics by humans, and evolve resistance to them.
Though antibiotics save countless lives every year, they must also be used carefully to avoid the further spread of antibiotic resistant bacteria and diseases. Better understanding how bacteria evolve resistance will allow us to improve how we use our current antibiotics, as well as the ones we will develop in the future.
ABOUT THE AUTHOR
Celia Souque is a Postdoctoral Researcher, Microbiology, University of Oxford. Celia is a microbiologist fascinated by the challenge created by antibiotic resistance (AMR) and the multiplicity of approaches needed to tackle it. After a PhD focusing on unraveling the evolutionary dynamics of an important driver of antimicrobial resistance at the University of Oxford she is now working on the impact of the colistin ban in agriculture on resistance in China in the same university. Celia has a keen interest in developing novel and effective ways of communicating around the problem of AMR, both to the public and to policy makers. During her PhD, she created a boardgame about antibiotic resistance for the Thai public, now available as a free educational resource online.
This article is courtesy of The Conversation.